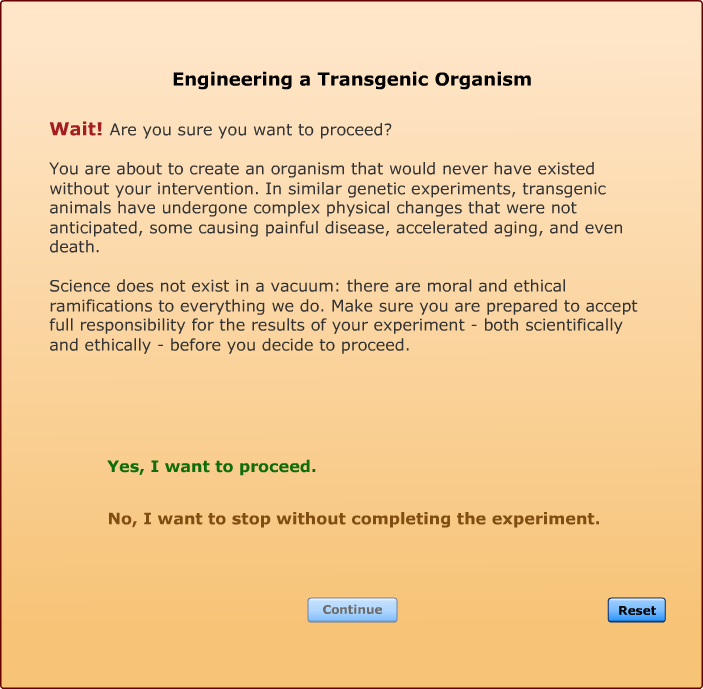
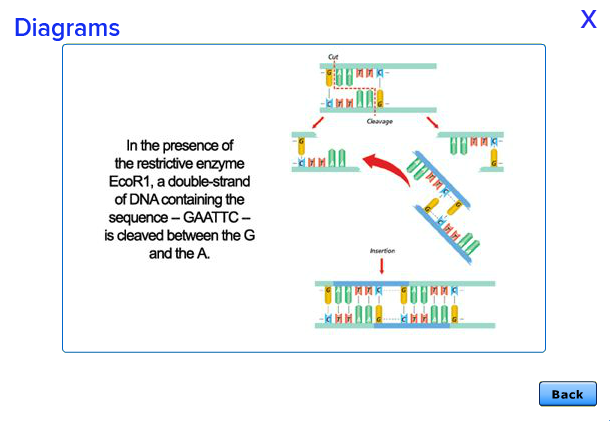
In this Lab, you will take the fragment of DNA that causes a unique trait in one organism such as the DNA that makes a firefly glow and splice that DNA into a different organism, so that the second organism takes on the trait of the first organism. At least, that's what you will attempt to do. The results may not always be what you expect. In the controversial field of genetic engineering, heated claims and passionate counterclaims are everywhere. And as technology advances, both the promises and the objections are rapidly multiplying.
We have designed a gene splicing simulator that follows many of the same steps that bioengineers use when they create transgenic organisms. By using the simulator, you will increase your understanding of what is involved in gene splicing, you will learn the major technical problems that must be solved, you will master the different procedures used, and you will study some of the potential dangers and ethical questions associated with bioengineering.
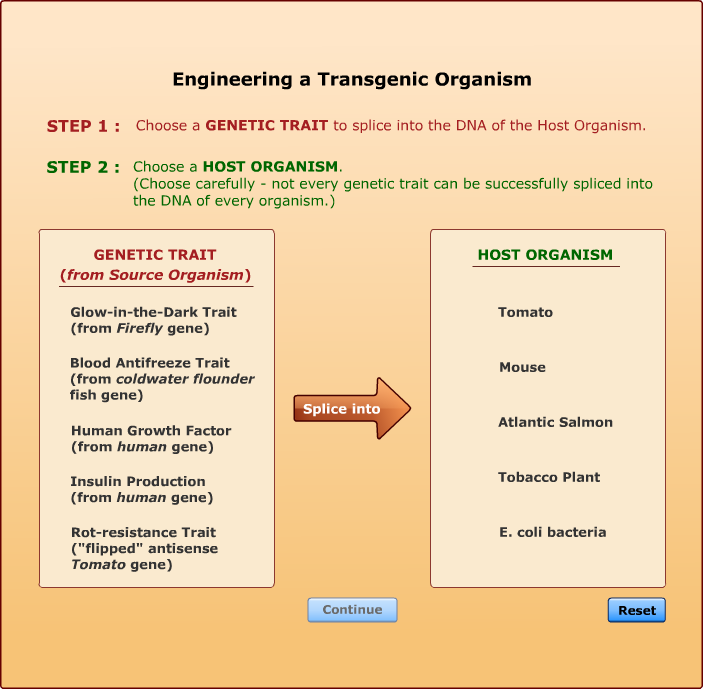
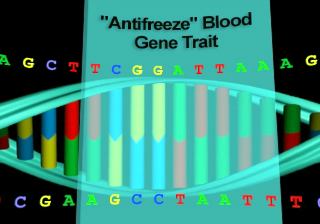
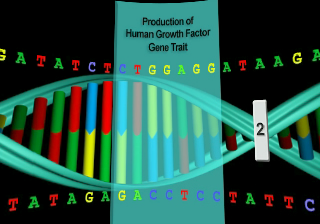
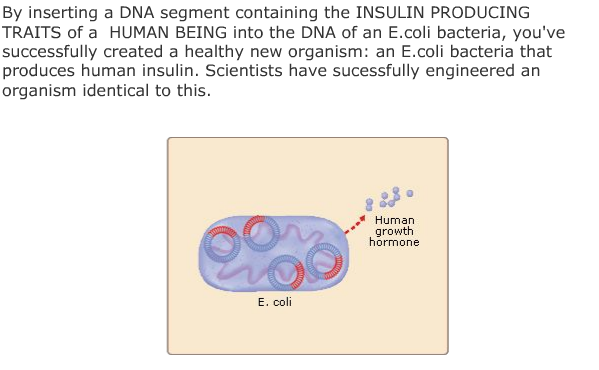
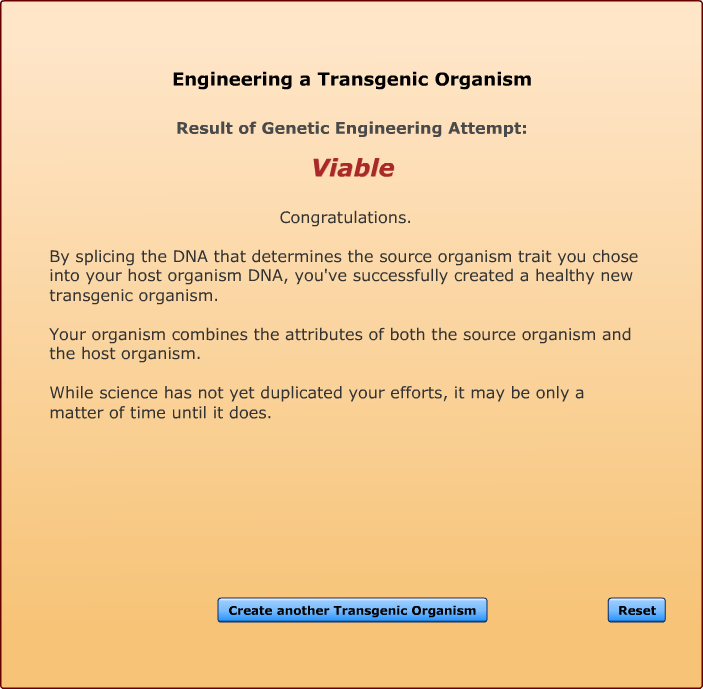
All of the genetic traits that appear in the gene attribute list on this screen have been spliced into one of the listed host organisms by bioengineers. Each has resulted in the creation of a viable transgenic organism. Not every transgenic animal you engineer will be able to survive in the real world, but if you keep trying, you will soon end up with a unique creature, one that was produced from the DNA of two completely different organisms.
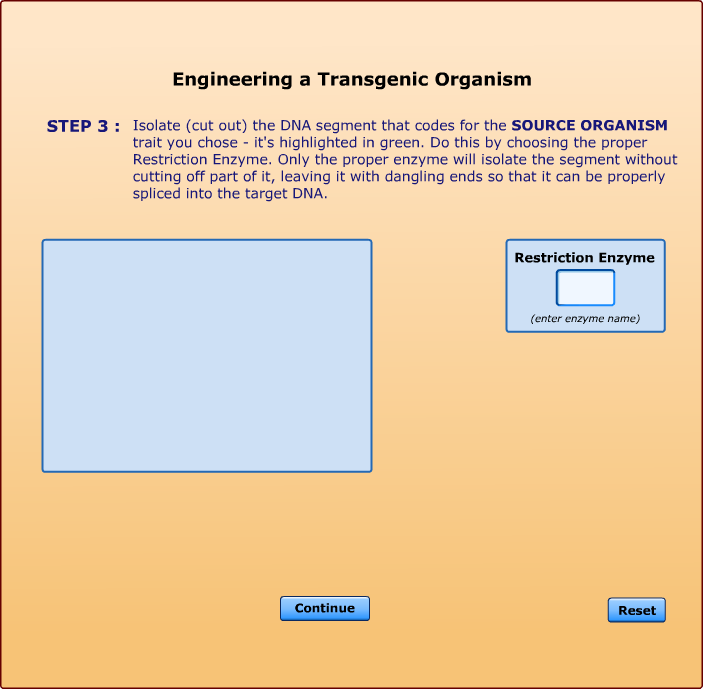
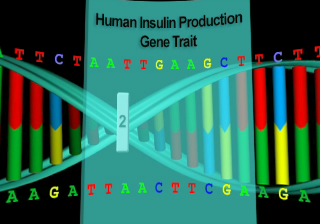
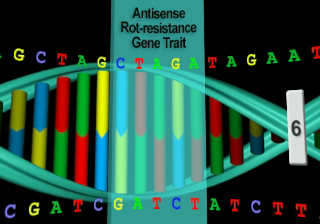






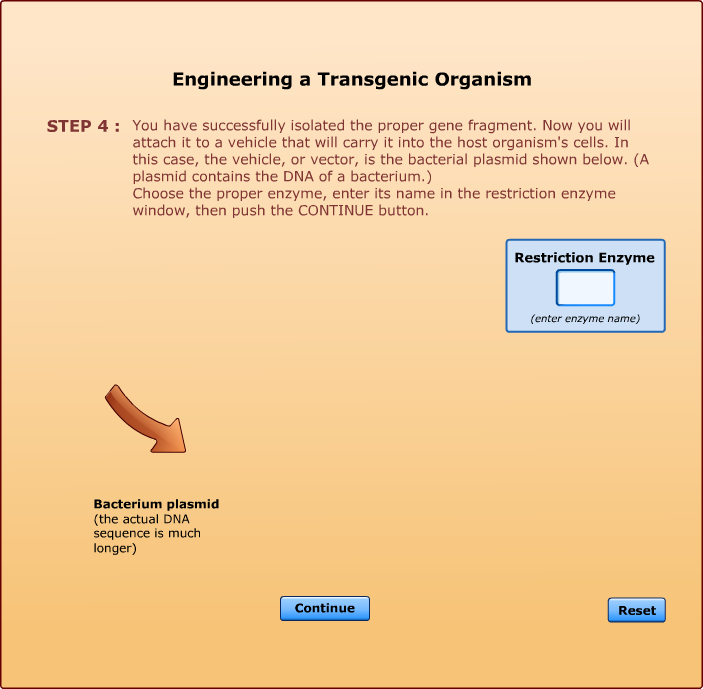
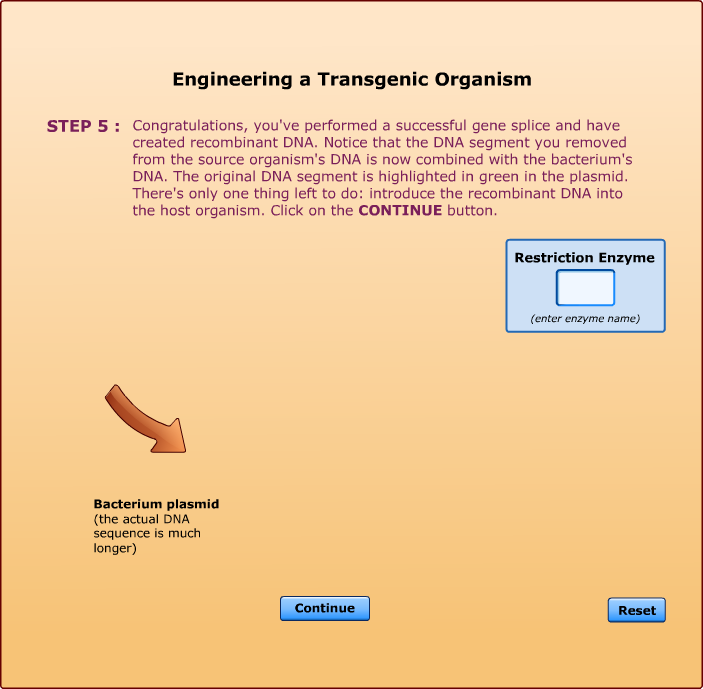









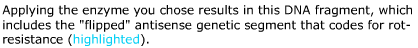
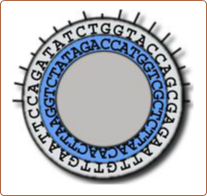
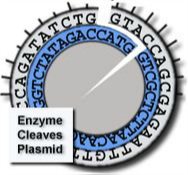
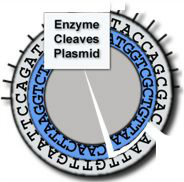
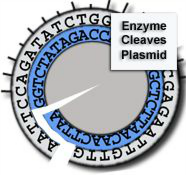
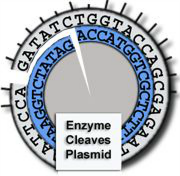
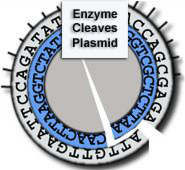
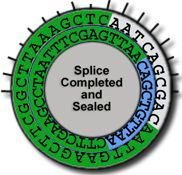
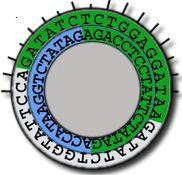































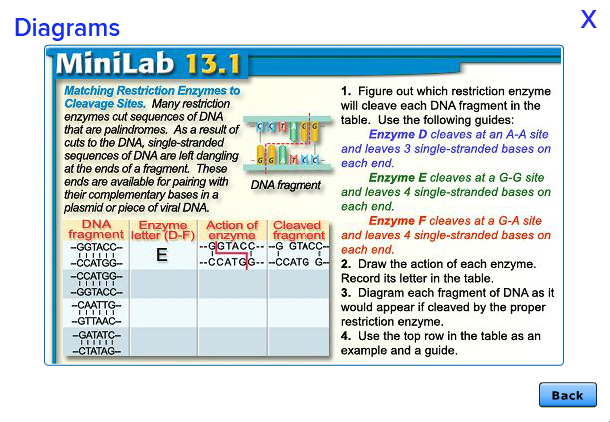





Cleaves at an A-A site -
leaves 4 single-stranded
bases on each end.
Enzyme B:
Cleaves at a G-A site -
leaves 4 single-stranded
bases on each end.
Enzyme C:
Cleaves at a G-C site -
cleaves with a blunt cut,
leaving no sticky ends.
Enzyme D:
Cleaves at a C-A site -
leaves 4 single-stranded
bases on each end.
Enzyme E:
Cleaves at a G-G site -
leaves 4 single-stranded
bases on each end.
Enzyme F:
Cleaves at a G-A site -
leaves 4 single-stranded
bases on each end.
Enzyme G:
Cleaves at a C-G site -
cleaves with a blunt cut,
leaving no sticky ends.
Enzyme H:
Cleaves at a A-G site -
leaves 6 single-stranded
bases on each end.
Enzyme I:
Cleaves at a C-G site -
leaves 6 single-stranded
bases on each end.
Enzyme J:
Cleaves at a A-A site - leaves 4 single-stranded
bases on each end.